Eclust Vignette
Load Required Packages
library(knitr)
library(data.table)
library(magrittr)
library(ggplot2)
library(latex2exp)
library(dplyr)
library(plyr)
library(glmnet)
library(stringr)
library(DT)
library(progress)
Simulate Data
set.seed(123456)
# number of predictors
p = 10
# number of test subjects
n = 200
# correlation between X's
rho = 0.50
# signal to noise ratio
signal_to_noise_ratio = 4
# names of the main effects, this will be used in many of the functions
main_effect_names <- paste0("x",1:p)
# names of the active set
true_var_names <- c("x1","x2","x3","x4","x1:x2", "x1:x3", "x1:x4", "x2:x3", "x2:x4", "x3:x4")
# different true coefficient vectors as in Table 1 of Choi et al.
beta1 <- c(7,2,1,1,0,0,0,0,0,0) %>% magrittr::set_names(true_var_names)
beta2 <- c(7,2,1,1,1,0,0,0.5,0.4,0.1) %>% magrittr::set_names(true_var_names)
beta3 <- c(7,2,1,1,7,7,7,2,2,1) %>% magrittr::set_names(true_var_names)
beta4 <- c(7,2,1,1,14,14,14,4,4,2) %>% magrittr::set_names(true_var_names)
beta5 <- c(0,0,0,0,7,7,7,2,2,1) %>% magrittr::set_names(true_var_names)
# simulate Toeplitz like correlation structure between X's
H <- abs(outer(1:p, 1:p, "-"))
cor <- rho^H
# generate X's from multivariate normal and label the matrix
DT <- MASS::mvrnorm(n = n, mu = rep(0,p), Sigma = cor) %>%
magrittr::set_colnames(paste0("x",1:p)) %>%
set_rownames(paste0("Subject",1:n))
# create X matrix which contains all main effects and interactions
# but not the intercept
X <- model.matrix(
as.formula(paste0("~(",paste0(main_effect_names, collapse = "+"),")^2-1")),
data = DT %>% as.data.frame())
# generate response with user defined signal to noise ratio
y.star <- X[,names(beta1)] %*% beta1
error <- rnorm(n)
k <- sqrt(var(y.star)/(signal_to_noise_ratio*var(error)))
Y <- y.star + k*error
colnames(Y) <- "Y"
# names of interaction variables assuming interaction terms contain a ":"
# this will be used in many of the functions
# names must appear in the same order as X matrix
interaction_names <- grep(":", colnames(X), value = T)
main_effect_names <- setdiff(colnames(X), interaction_names)
Analysis
Running the Strong heredity interaction model once using the shim function
NOTE: if lambda.beta = NULL and lambda.gamma = NULL then this function will use a grid of tuning parameters based on nlambda.beta only and the nlambda.gamma parameter is ignored. Therefore, if for example nlambda.beta = 10 then you must specify nlambda = 10x10 = 100 otherwise the function will break.
# load eclust library
library(eclust)
res <- shim(x = X, y = Y,
main.effect.names = main_effect_names,
interaction.names = interaction_names,
verbose = FALSE)
names(res)
## [1] "b0" "beta" "alpha"
## [4] "gamma" "lambda.beta" "lambda.gamma"
## [7] "tuning.parameters" "dfbeta" "dfalpha"
## [10] "dev.ratio" "deviance" "converged"
## [13] "x" "y" "bx"
## [16] "by" "sx" "center"
## [19] "normalize" "nlambda.gamma" "nlambda.beta"
## [22] "nlambda" "interaction.names" "main.effect.names"
## [25] "call" "nobs"
res
##
## Call: shim(x = X, y = Y, main.effect.names = main_effect_names, interaction.names = interaction_names, verbose = FALSE)
##
## dfBeta dfAlpha percentDev LambdaBeta LambdaGamma
## s1 8 28 0.8485 8.304e-06 8.304e-06
## s2 7 21 0.8337 8.304e-06 3.854e-05
## s3 7 21 0.8337 8.304e-06 1.789e-04
## s4 7 21 0.8336 8.304e-06 8.304e-04
## s5 7 21 0.8333 8.304e-06 3.854e-03
## s6 7 19 0.8323 8.304e-06 1.789e-02
## s7 7 16 0.8283 8.304e-06 8.304e-02
## s8 7 11 0.8239 8.304e-06 3.854e-01
## s9 7 6 0.8170 8.304e-06 1.789e+00
## s10 6 5 0.8134 8.304e-06 8.304e+00
## s11 6 15 0.8288 3.854e-05 8.304e-06
## s12 7 21 0.8345 3.854e-05 3.854e-05
## s13 7 21 0.8337 3.854e-05 1.789e-04
## s14 7 21 0.8336 3.854e-05 8.304e-04
## s15 7 21 0.8333 3.854e-05 3.854e-03
## s16 7 19 0.8323 3.854e-05 1.789e-02
## s17 7 16 0.8283 3.854e-05 8.304e-02
## s18 7 11 0.8239 3.854e-05 3.854e-01
## s19 6 6 0.8164 3.854e-05 1.789e+00
## s20 6 5 0.8132 3.854e-05 8.304e+00
## s21 5 10 0.8215 1.789e-04 8.304e-06
## s22 7 21 0.8338 1.789e-04 3.854e-05
## s23 7 21 0.8338 1.789e-04 1.789e-04
## s24 7 21 0.8337 1.789e-04 8.304e-04
## s25 7 21 0.8334 1.789e-04 3.854e-03
## s26 7 19 0.8323 1.789e-04 1.789e-02
## s27 7 16 0.8284 1.789e-04 8.304e-02
## s28 6 9 0.8177 1.789e-04 3.854e-01
## s29 5 3 0.8079 1.789e-04 1.789e+00
## s30 5 2 0.8047 1.789e-04 8.304e+00
## s31 5 10 0.8139 8.304e-04 8.304e-06
## s32 5 10 0.8172 8.304e-04 3.854e-05
## s33 5 10 0.8172 8.304e-04 1.789e-04
## s34 5 10 0.8172 8.304e-04 8.304e-04
## s35 5 10 0.8170 8.304e-04 3.854e-03
## s36 5 10 0.8163 8.304e-04 1.789e-02
## s37 5 9 0.8144 8.304e-04 8.304e-02
## s38 5 6 0.8100 8.304e-04 3.854e-01
## s39 4 2 0.8044 8.304e-04 1.789e+00
## s40 4 1 0.8042 8.304e-04 8.304e+00
## s41 3 3 0.8043 3.854e-03 8.304e-06
## s42 3 3 0.8042 3.854e-03 3.854e-05
## s43 3 3 0.8042 3.854e-03 1.789e-04
## s44 3 3 0.8042 3.854e-03 8.304e-04
## s45 3 3 0.8042 3.854e-03 3.854e-03
## s46 3 3 0.8042 3.854e-03 1.789e-02
## s47 3 3 0.8043 3.854e-03 8.304e-02
## s48 3 3 0.8043 3.854e-03 3.854e-01
## s49 3 3 0.8041 3.854e-03 1.789e+00
## s50 3 1 0.8038 3.854e-03 8.304e+00
## s51 3 3 0.8043 1.789e-02 8.304e-06
## s52 3 3 0.8043 1.789e-02 3.854e-05
## s53 3 3 0.8043 1.789e-02 1.789e-04
## s54 3 3 0.8043 1.789e-02 8.304e-04
## s55 3 3 0.8043 1.789e-02 3.854e-03
## s56 3 3 0.8043 1.789e-02 1.789e-02
## s57 3 3 0.8043 1.789e-02 8.304e-02
## s58 3 3 0.8043 1.789e-02 3.854e-01
## s59 3 3 0.8041 1.789e-02 1.789e+00
## s60 3 1 0.8038 1.789e-02 8.304e+00
## s61 3 3 0.8043 8.304e-02 8.304e-06
## s62 3 3 0.8043 8.304e-02 3.854e-05
## s63 3 3 0.8043 8.304e-02 1.789e-04
## s64 3 3 0.8043 8.304e-02 8.304e-04
## s65 3 3 0.8043 8.304e-02 3.854e-03
## s66 3 3 0.8043 8.304e-02 1.789e-02
## s67 3 3 0.8043 8.304e-02 8.304e-02
## s68 3 3 0.8043 8.304e-02 3.854e-01
## s69 3 2 0.8041 8.304e-02 1.789e+00
## s70 3 1 0.8039 8.304e-02 8.304e+00
## s71 3 3 0.8037 3.854e-01 8.304e-06
## s72 3 3 0.8037 3.854e-01 3.854e-05
## s73 3 3 0.8036 3.854e-01 1.789e-04
## s74 3 3 0.8036 3.854e-01 8.304e-04
## s75 3 3 0.8036 3.854e-01 3.854e-03
## s76 3 3 0.8036 3.854e-01 1.789e-02
## s77 3 3 0.8036 3.854e-01 8.304e-02
## s78 3 3 0.8036 3.854e-01 3.854e-01
## s79 3 2 0.8034 3.854e-01 1.789e+00
## s80 3 1 0.8032 3.854e-01 8.304e+00
## s81 1 0 0.7170 1.789e+00 8.304e-06
## s82 1 0 0.7170 1.789e+00 3.854e-05
## s83 1 0 0.7170 1.789e+00 1.789e-04
## s84 1 0 0.7170 1.789e+00 8.304e-04
## s85 1 0 0.7170 1.789e+00 3.854e-03
## s86 1 0 0.7170 1.789e+00 1.789e-02
## s87 1 0 0.7170 1.789e+00 8.304e-02
## s88 1 0 0.7170 1.789e+00 3.854e-01
## s89 1 0 0.7170 1.789e+00 1.789e+00
## s90 1 0 0.7170 1.789e+00 8.304e+00
## s91 1 0 0.7035 8.304e+00 8.304e-06
## s92 1 0 0.7035 8.304e+00 3.854e-05
## s93 1 0 0.7035 8.304e+00 1.789e-04
## s94 1 0 0.7035 8.304e+00 8.304e-04
## s95 1 0 0.7035 8.304e+00 3.854e-03
## s96 1 0 0.7035 8.304e+00 1.789e-02
## s97 1 0 0.7035 8.304e+00 8.304e-02
## s98 1 0 0.7035 8.304e+00 3.854e-01
## s99 1 0 0.7035 8.304e+00 1.789e+00
## s100 1 0 0.7035 8.304e+00 8.304e+00
plot(res)
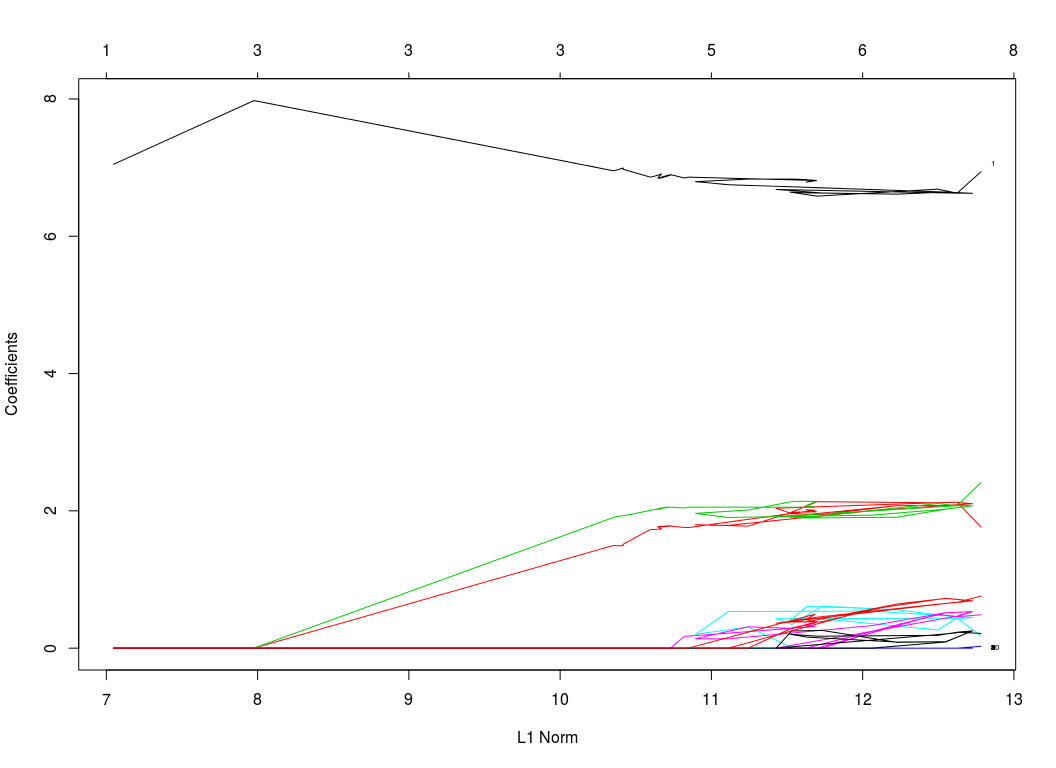
Cross Validation using the cv.shim function
library(doMC)
registerDoMC(cores = 4)
cv.res <- cv.shim(x = X, y = Y,
main.effect.names = main_effect_names,
interaction.names = interaction_names,
parallel = TRUE, verbose = FALSE,
type.measure = c("mse"),
nfolds = 5)
names(cv.res)
## [1] "lambda.beta" "lambda.gamma" "cvm"
## [4] "cvsd" "cvup" "cvlo"
## [7] "nz.main" "name" "nz.interaction"
## [10] "shim.fit" "converged" "cvm.mat.all"
## [13] "df" "lambda.min.beta" "lambda.min.name"
## [16] "lambda.1se.beta" "lambda.1se.name"
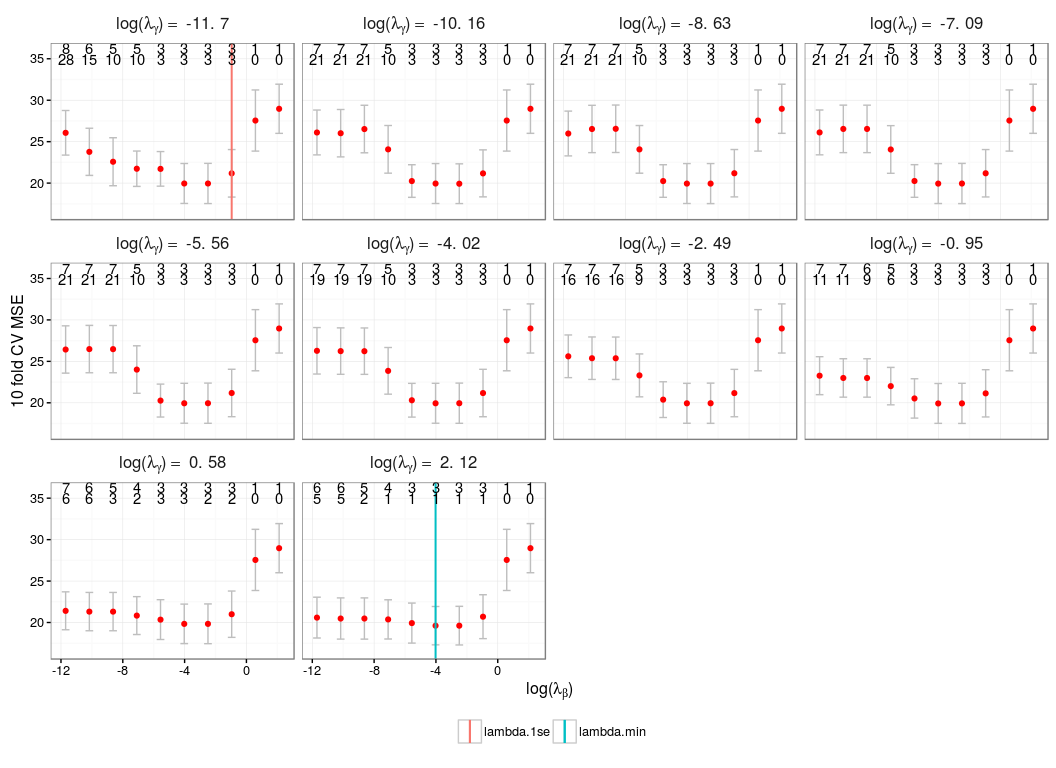
Cross Validation Plot
plot(cv.res)
Coefficient Estimates
coef(cv.res, s = "lambda.1se")
## 56 x 1 sparse Matrix of class "dgCMatrix"
## Y
## X s71
## (Intercept) -0.07660618
## x1 6.97690913
## x2 1.51190024
## x3 1.92657143
## x4 .
## x5 .
## x6 .
## x7 .
## x8 .
## x9 .
## x10 .
## x1:x2 -0.06471414
## x1:x3 -0.19559520
## x1:x4 .
## x1:x5 .
## x1:x6 .
## x1:x7 .
## x1:x8 .
## x1:x9 .
## x1:x10 .
## x2:x3 0.63248399
## x2:x4 .
## x2:x5 .
## x2:x6 .
## x2:x7 .
## x2:x8 .
## x2:x9 .
## x2:x10 .
## x3:x4 .
## x3:x5 .
## x3:x6 .
## x3:x7 .
## x3:x8 .
## x3:x9 .
## x3:x10 .
## x4:x5 .
## x4:x6 .
## x4:x7 .
## x4:x8 .
## x4:x9 .
## x4:x10 .
## x5:x6 .
## x5:x7 .
## x5:x8 .
## x5:x9 .
## x5:x10 .
## x6:x7 .
## x6:x8 .
## x6:x9 .
## x6:x10 .
## x7:x8 .
## x7:x9 .
## x7:x10 .
## x8:x9 .
## x8:x10 .
## x9:x10 .
coef(cv.res, s = "lambda.min")
## 56 x 1 sparse Matrix of class "dgCMatrix"
## Y
## X s60
## (Intercept) -0.1499439
## x1 6.8466272
## x2 1.7668964
## x3 2.0307660
## x4 .
## x5 .
## x6 .
## x7 .
## x8 .
## x9 .
## x10 .
## x1:x2 .
## x1:x3 .
## x1:x4 .
## x1:x5 .
## x1:x6 .
## x1:x7 .
## x1:x8 .
## x1:x9 .
## x1:x10 .
## x2:x3 0.5483146
## x2:x4 .
## x2:x5 .
## x2:x6 .
## x2:x7 .
## x2:x8 .
## x2:x9 .
## x2:x10 .
## x3:x4 .
## x3:x5 .
## x3:x6 .
## x3:x7 .
## x3:x8 .
## x3:x9 .
## x3:x10 .
## x4:x5 .
## x4:x6 .
## x4:x7 .
## x4:x8 .
## x4:x9 .
## x4:x10 .
## x5:x6 .
## x5:x7 .
## x5:x8 .
## x5:x9 .
## x5:x10 .
## x6:x7 .
## x6:x8 .
## x6:x9 .
## x6:x10 .
## x7:x8 .
## x7:x9 .
## x7:x10 .
## x8:x9 .
## x8:x10 .
## x9:x10 .