Gene by environment interactions
High-dimensional gene by environment interactions with applications to maternal and child health
High-dimensional (HD) genomic data \((\mathbf{X})\) often reflect a tissue’s functioning and can be strongly influenced by lifestyle or demographic exposures \((E)\), making the interactions \((\mathbf{X} \cdot E)\) crucial for predicting outcomes \((Y)\). However, the sheer number of potential interactions, their non-linear nature, and the low power to detect them pose major challenges. In precision medicine, where the goal is to tailor interventions to individual risk profiles, it is equally important to build accurate models and to uncover which features—among possibly correlated or latent factors—actually drive predictions. Recent evidence also suggests that environmental exposures can broadly alter regulatory networks and that assessing gene similarity or co-regulation can capture these systemic effects better than simply modeling raw expression data. Yet most existing predictive methods that use HD data and exposures struggle with low power, limited interpretability, and difficulty handling interactions, underscoring the need for more appropriate analytic approaches.
To address this gap, we propose ECLUST (Bhatnagar et al., 2018), a conceptual analytic strategy that uses exposure-sensitive data clusters in two-step algorithms for continuous or binary outcome prediction in HD contexts. We hypothesize that incorporating exposure data into variable grouping can improve both predictive accuracy and interpretability. Our approach is motivated by maternal-child health research, where events during pregnancy—such as gestational diabetes mellitus (GDM)—are suspected to influence the risk of childhood obesity. Indeed, children born to women with GDM are more likely to be overweight or obese, and epigenetic factors have emerged as crucial mediators. Methylation changes in placenta and cord blood have been linked to GDM, and here we explore how these changes relate to childhood obesity outcomes at around five years of age. By grouping features that exhibit a systematic response to GDM, ECLUST retains a systems-based perspective and avoids focusing solely on single markers. Through simulations and real data analyses, this approach has demonstrated improved prediction accuracy and interpretability in both linear and non-linear settings, shedding light on how exposures like GDM reshape genomic co-regulation and ultimately influence childhood obesity risk. An R package implementing ECLUST is available on CRAN, making it accessible to a wide range of researchers and practitioners.
Building on this foundation of exposure-centric modeling, we developped a new penalized regression method called SAIL that specifically targets non-linear interactions between a single key exposure and a large set of features under strong or weak heredity constraints (Bhatnagar et al., 2023). This means that an interaction term is included in the model only if the corresponding main effects are also present (strong) or at least one of them is present (weak). Importantly, SAIL is shown to possess the oracle property asymptotically, implying that it can recover the true underlying model as well as if that model were known in advance. From a practical standpoint, SAIL employs a computationally efficient fitting algorithm with automatic selection of tuning parameters, allowing it to scale well to modern HD datasets. Simulation studies demonstrate its superiority over other penalized methods in accurately predicting outcomes and recovering the true set of active (and interacting) variables when interactions are genuinely non-linear. The proposed algorithms are implemented in an R package available on GitHub.
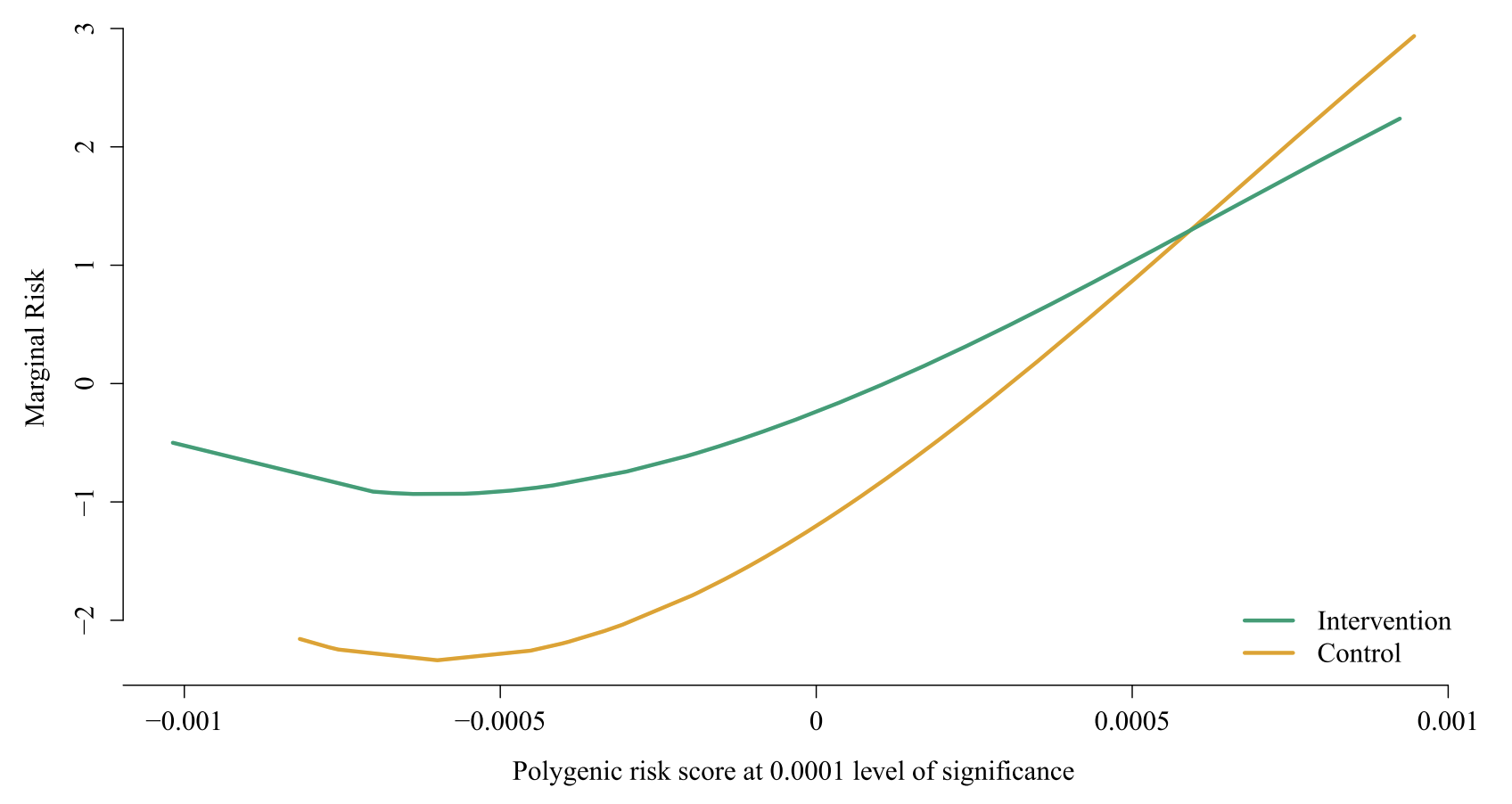
A compelling example of SAIL’s utility comes from a study of the Nurse Family Partnership (NFP) program. NFP is a psychosocial intervention program targeting low-income mothers with the goal of improving pregnancy outcomes, children’s health and development, and long-term economic self-sufficiency. Beginning in pregnancy and continuing through infancy, mothers receive regular home visits from nurses who provide guidance on maternal health, parenting, and mother-infant interactions. In a randomized trial, pregnant women were assigned either to the nurse-visited group (intervention) or a control group, and child IQ (using Stanford Binet scores) was measured at four years of age. In this scenario, researchers were interested in how a child’s genetic risk for educational attainment—summarized by a polygenic risk score (PRS)—might interact with participation in NFP to influence IQ at age four. Applying SAIL revealed that children with lower PRS scores (i.e., those genetically predisposed to lower educational attainment) benefited most from the intervention, indicating a clear gene-environment interaction (see Figure 2).
A third advancement expands hierarchical gene-environment interaction (GEI) modeling to high-dimensional mixed models. A longstanding challenge in GEI analysis is that population structure and closer relatedness, alongside shared environmental exposures, can create correlations that lead to spurious signals if not properly modeled. Recognizing that ignoring these dependencies can inflate false positive rates under polygenic models, my PhD student, Julien, developed a penalized quasi-likelihood approach for hierarchical variable selection within generalized linear mixed models (GLMMs) called pglmm (St-Pierre et al., 2024). By incorporating random effects to account for both genetic and environmental correlations, pglmm reduces false discoveries for main and interaction effects and achieves higher F1 scores than existing methods in simulation studies. The method is implemented in an open source Julia package.
A motivating application of pglmm centers on the Orofacial Pain: Prospective Evaluation and Risk Assessment (OPPERA) study, which previously reported significant associations between temporomandibular disorder (TMD)—a painful jaw condition disproportionately affecting females—and four distinct genetic loci. Because TMD may involve sex-specific pathophysiological mechanisms, pglmm was used to identify important sex-specific predictors in the OPPERA discovery cohort and then evaluate the model’s predictive performance in two replication cohorts: OPPERA II Chronic TMD Replication and Complex Persistent Pain Conditions (CPPC). pglmm successfully retrieved the previously implicated loci, highlighting its ability to detect biologically relevant interactions and confirm risk predictions in independent samples. These results underscore the importance of accounting for correlated structures in GEI analyses and demonstrate how hierarchical selection in a mixed-model framework can improve both discovery and replication in complex trait genetics.