Polygenic risk scores
Polygenic risk scores for complex diseases with applications in screening, risk stratification, and identification of rare variants
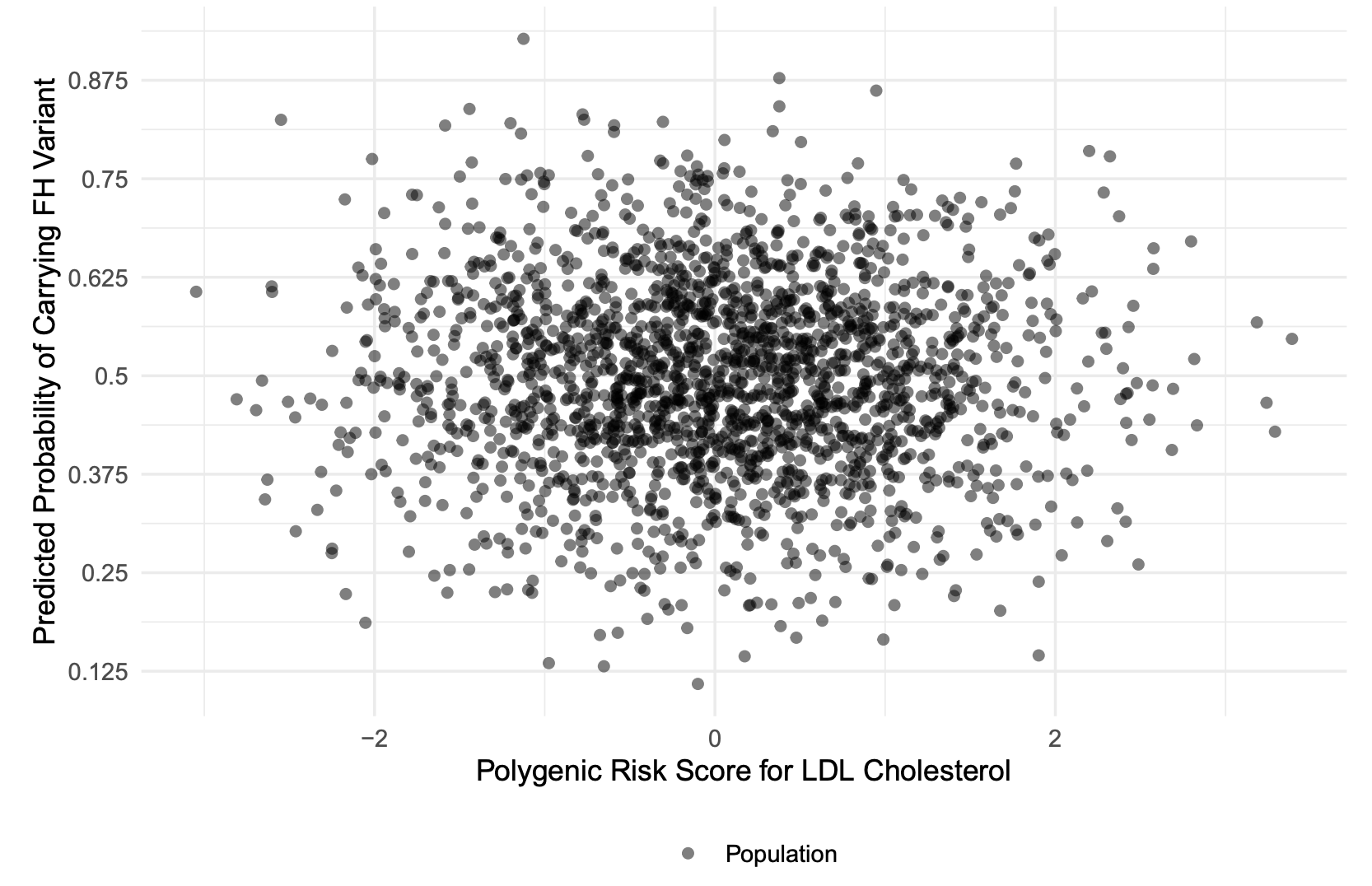
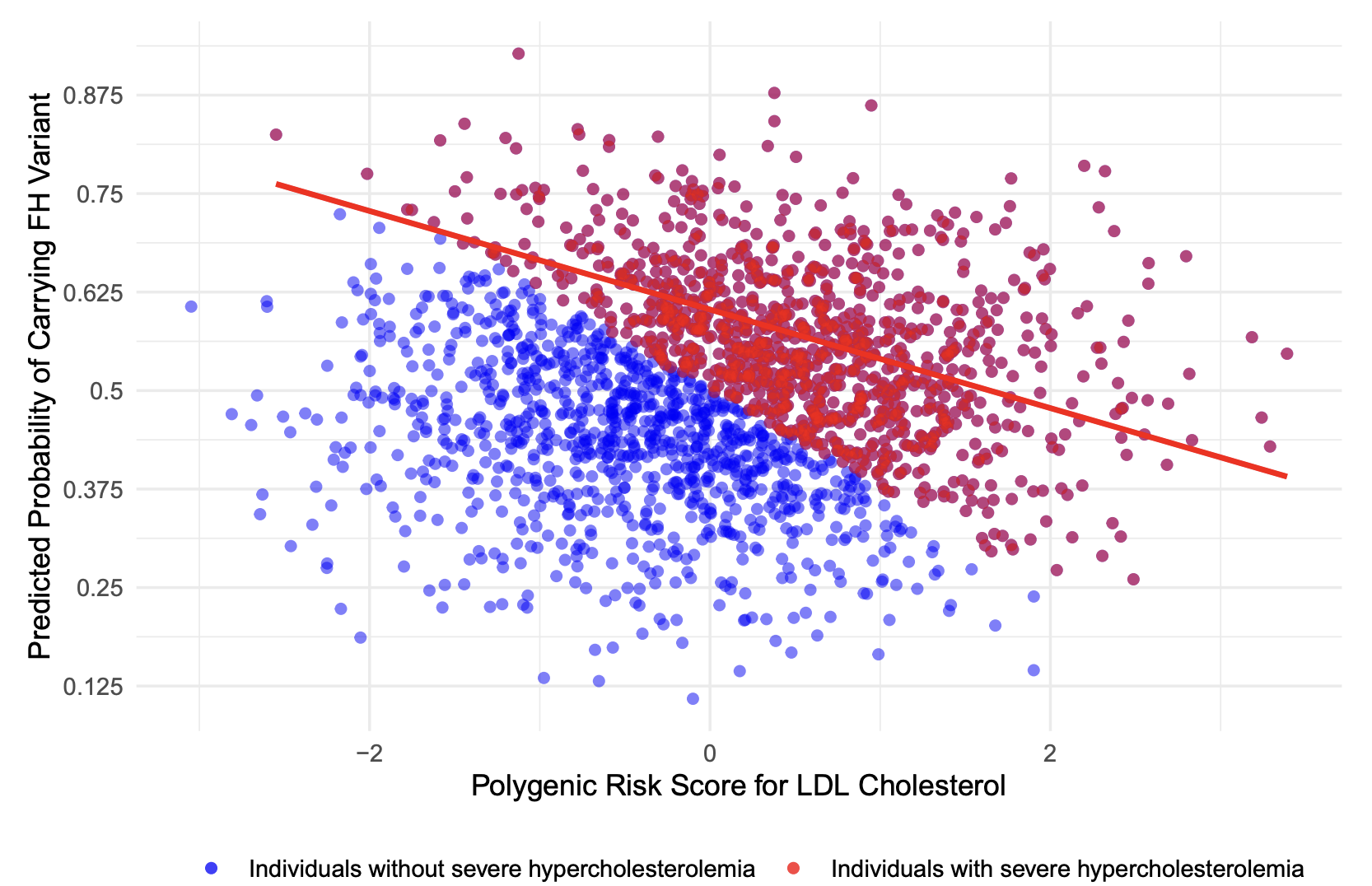
We have been developing new polygenic risk score (PRS) methods for complex diseases to improve screening, risk stratification, and the identification of individuals with rare variant carriers. In a study led by my Master’s student, Haoyu Wu (co-supervised by Dr. Brent Richards), we investigated the clinical potential of a low-density lipoprotein cholesterol (LDL-C) PRS to identify carriers of Familial Hypercholesterolemia (FH)—a disorder that often warrants costly therapies, such as PCSK9 inhibitors, to prevent cardiovascular outcomes (Wu et al., 2021). Because routine genetic testing for FH among hypercholesterolemia patients remains expensive and administratively challenging, we used a relatively inexpensive genome-wide genotyping strategy to derive an LDL-C PRS. Our approach explained 21% of the variance in LDL-C levels in White British individuals from the UK Biobank. Notably, among patients with severe hypercholesterolemia, those with a low LDL-C PRS were 21 times more likely to harbor an FH variant compared to those with a high PRS—a difference attributable in part to collider bias (Figure 1). In this context, collider bias arises because we conditioned on the common outcome of severe hypercholesterolemia, which can result from either a rare FH pathogenic variant or a polygenic predisposition captured by the PRS. Consequently, once we focused on individuals who developed severe hypercholesterolemia, those with a low polygenic risk stood out as more likely to carry the monogenic cause. By leveraging this negative correlation between PRS and FH variants in the presence of severe hypercholesterolemia, PRS-based triaging emerges as a viable, cost-effective strategy for enhancing FH detection and prioritizing genetic testing.
We also investigated the use of PRSs as a negative screening tool. For example, we developed a PRS for quantitative ultrasound speed of sound (SOS) to improve screening for fracture risk (Forgetta et al., 2020; Lu et al., 2020). Osteoporosis is a common disease characterized by low bone mineral density and increased fracture risk. Screening for osteoporosis is typically done using the Fracture Risk Assessment Tool (FRAX), which calculates a 10-year probability of major osteoporotic fracture. However, current screening guidelines identify only a small proportion of the population as eligible for intervention, meaning that much of the screening expenditure is spent on individuals who will not qualify for intervention. In our study, we developed a PRS for SOS, a heritable risk factor for osteoporotic fracture. We found that this PRS could identify individuals at low risk of osteoporosis who could safely be excluded from further screening. This approach could potentially reduce the number of individuals needing to be screened, while maintaining a high sensitivity and specificity to identify individuals who should be recommended an intervention.
My work on polygenic risk scores (PRS) has also focused on overcoming key methodological hurdles in high-dimensional genetic prediction, including population structure, relatedness, and binary outcomes. First, we developped ggmix, which provides a single-step penalized linear mixed model that adjusts for both population structure and correlated SNPs in continuous traits; its efficient blockwise coordinate descent algorithm is freely available in an open-source R package (Bhatnagar et al., 2020). Building on this framework, my PhD student Julien St-Pierre (co-supervised with Dr. Karim Oualkacha) led the development of pglmm, which extends penalized mixed-model methods to generalized linear mixed models for binary traits (St-Pierre et al., 2023). By accounting for between-individual correlations via random effects, pglmm selects predictive markers more accurately than principal-component–adjusted methods in case-control studies; a Julia implementation supports large-scale analyses. Additionally, Julien also spearheaded a study on diverse SNP selection strategies for PRS, emphasizing how selection criteria affect predictive performance—especially when disease risk varies across populations (St-Pierre et al., 2022). Together, these advances strengthen the foundation for PRS research, as they address multiple sources of bias, improve power, and offer computationally efficient open-source solutions for practitioners in both research and clinical settings.