Machine Learning for survival analysis
Variable selection and absolute risk estimation for time-to-event outcomes using machine learning
Traditional survival analysis approaches like Cox regression focus on hazard ratios and often rely on time-matching or risk-set sampling, which removes the baseline hazard from the estimating equations. This makes it cumbersome to report or visualize absolute risks and survival curves because a second step is required to separately estimate the baseline hazard. To overcome these limitations, we developped the casebase framework (Bhatnagar et al., 2022), which extends the Hanley & Miettinen (2009) approach for fitting fully parametric hazard models and covariate-conditional survival curves using the familiar interface of the glm function. Our implementation includes extensions to other models such as penalized regression for variable selection and competing risk analysis. In addition, we provide functions for exploratory data analysis and visualizing the estimated quantities such as the hazard function, survival curve, and their standard errors. The ultimate goal of our package is to make fitting flexible hazards accessible to end users who favor reporting absolute risks and survival curves over hazard ratios. The package is available on CRAN and GitHub.
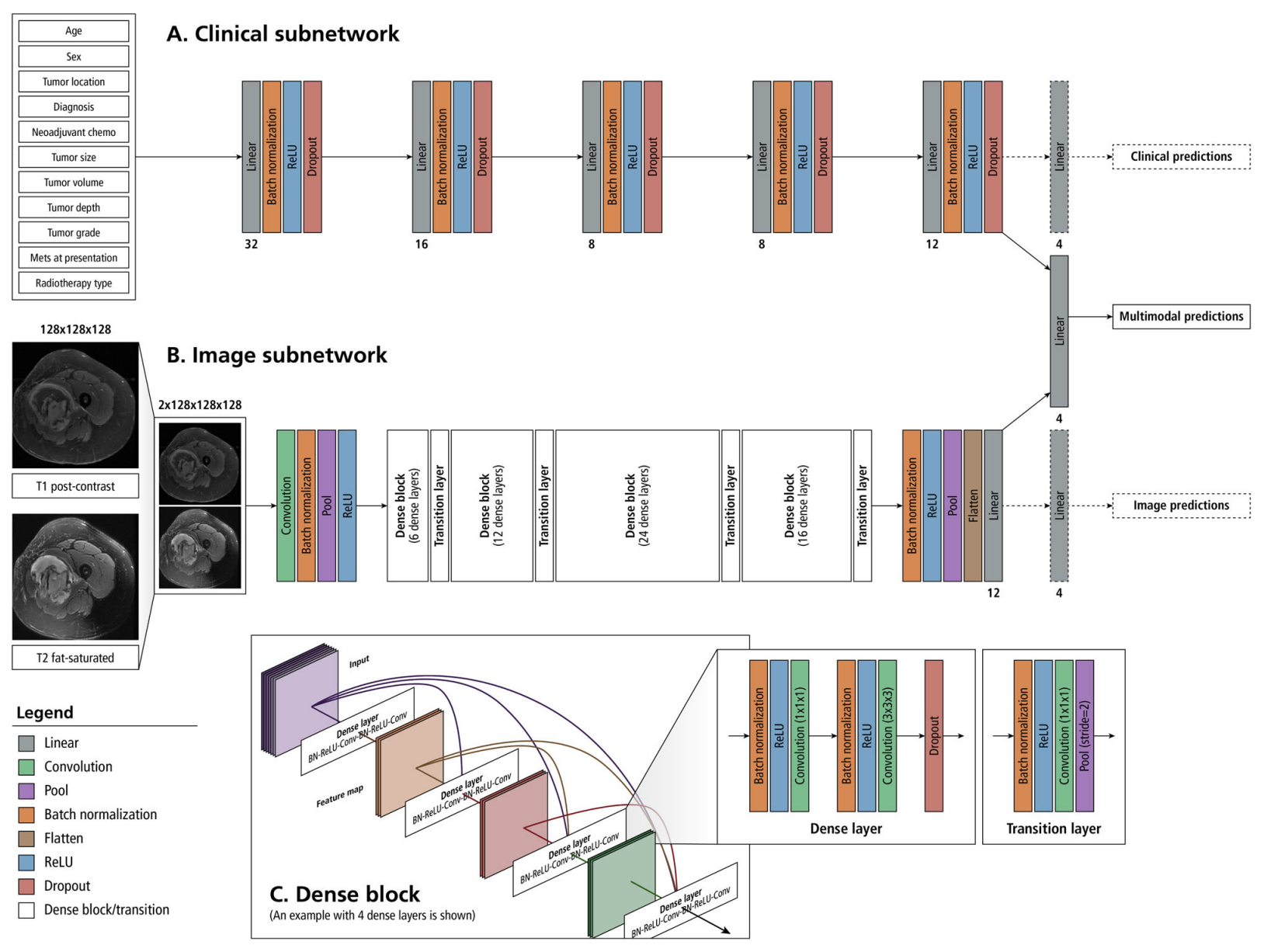
We applied casebase to the European Randomized Study of Prostate Cancer Screening (ERSPC) dataset to investigate the differences in prostate cancer risk between the control and screening arms. Previous re-analyses of these data suggest that the 20% reduction in prostate cancer death due to screening was an underestimate. The estimated 20% (from a proportional hazards model) did not account for the delay between screening and the time the effect is expected to be observed. As a result, the null effects in years 1–7 masked the substantial reductions that began to appear from year 8 onward. This motivates the use of a time-dependent hazard ratio which can easily be fit with the casebase package by including an interaction term with time in the model. We fit a flexible hazard by using a smooth function of time modeled with a penalized cubic spline basis with 2 degrees of freedom (implemented in the survival::pspline function). The resulting time-dependent hazard ratio is shown in Figure 1.
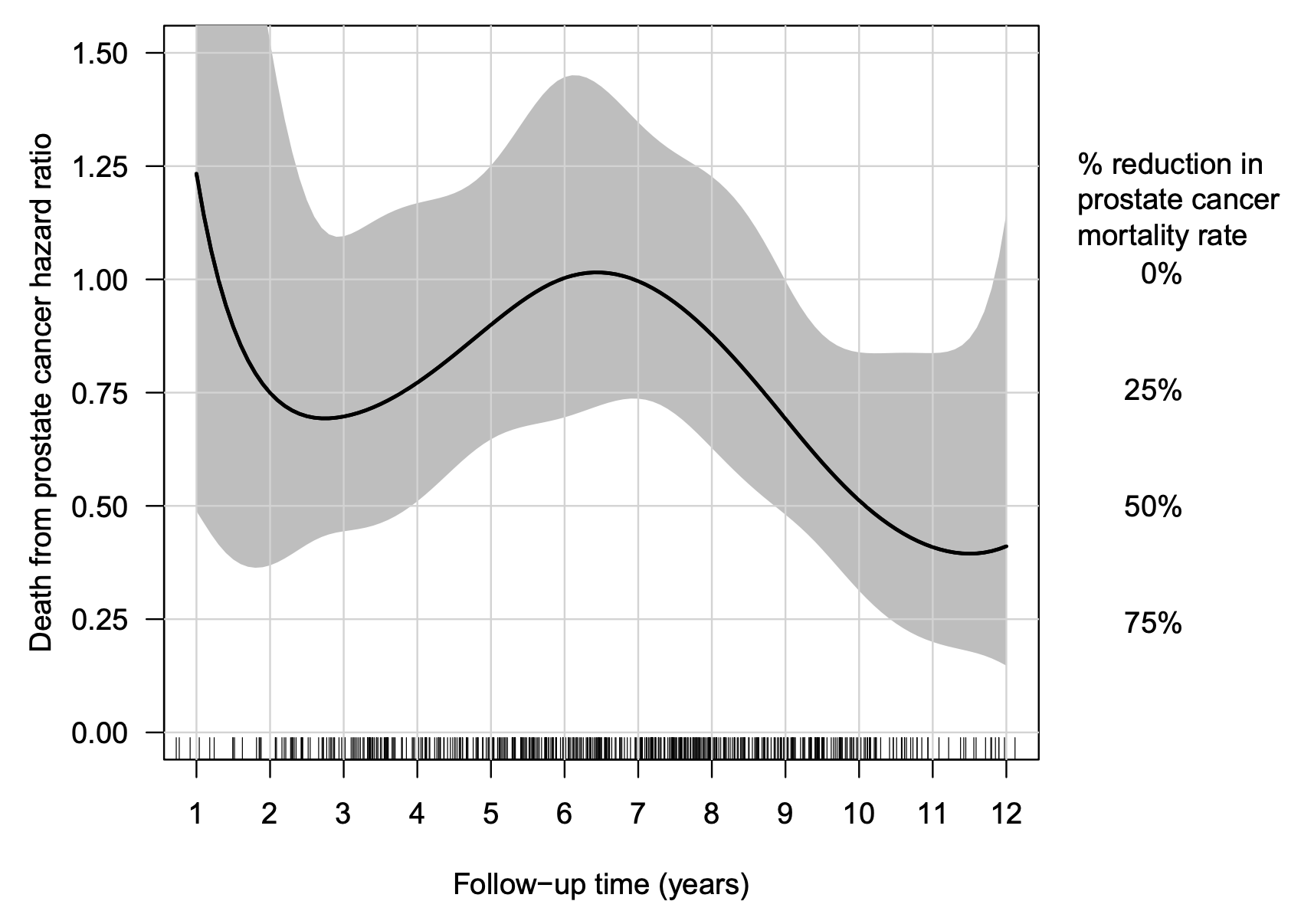
Recognizing that non-linear and high-order covariate interactions often underlie complex disease processes, my PhD student, Jesse Islam, led the development of Case-Base Neural Networks (CBNNs) (Islam et al., 2024) as a new approach that combines the case-base sampling framework with flexible neural network architectures. Using a novel sampling scheme and data augmentation to naturally account for censoring, we construct a feed-forward neural network that includes time as an input (Figure 2). Our results highlight the benefit of combining case-base sampling with deep learning to provide a simple and flexible framework for data-driven modeling of single event survival outcomes that estimates time-varying effects and a complex baseline hazard by design. An R package is available at https://github.com/Jesse-Islam/cbnn.
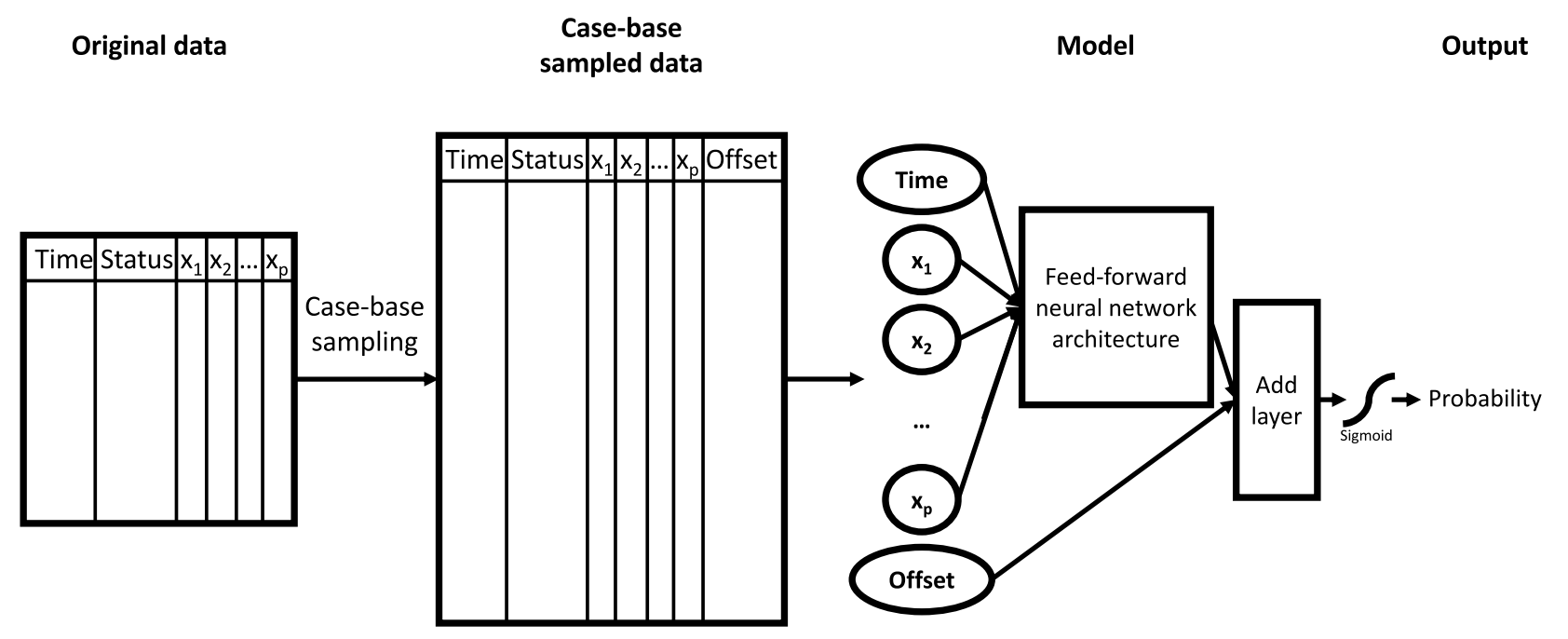
Beyond standard tabular data, imaging has become integral to many clinical prognoses. Led by my PhD student Anthony Bozzo, we built a multimodal neural network (MMNN) that processes both clinical variables and magnetic resonance imaging (MRI) slices from soft tissue sarcoma (STS) patients (Bozzo et al., 2024). By employing gradient blending, the network merges two sub-models—one handling 3D T1/T2 MRI volumes, the other handling clinical features—so that they can converge optimally without overfitting (Figure 3). Compared to unimodal and classical radiomics models, the MMNN exhibited superior performance for predicting overall survival and risk of distant metastases (C-index 0.77 and 0.70, respectively). Heat maps of salient image features provided clinically meaningful insights into regions of the tumor that drive the model’s predictions. This framework underscores the promise of end-to-end deep learning for integrating imaging and clinical data in complex survival tasks.