20 Packages
This section is adapted from The Epidemiologist R Handbook125
Packages are shareable collections of R code that can contain functions, data, and/or documentation. Packages increase the functionality of R by providing access to additional functions to suit a variety of needs. While it is entirely possible to do all of your work in R without ever using a package, we do not recommend that approach. There are a wealth of packages available, almost all of which help reduce both the learning curve associated with R and the amount of time spent on any given analytical project.
20.1 Installing a package with install.packages (not recommended)
In order to access the functions within a package, you must first install the package on your computer. There are a collection of R packages hosted on the internet on the CRAN website: CRAN(https://cran.r-project.org/), the Comprehensive R Archive Network. These packages must meet certain quality standards, and they are regularly tested.
If an R user feels that their package would benefit a broad audience, they may choose to submit their package to CRAN. The process of submitting a package and having it published through CRAN is beyond the scope of this Chapter, but it’s important to point out that you can create a package that you use all for yourself, share with colleagues, or submit to CRAN. Most of the packages we’ll be working with in this book are available on CRAN, which means that we can install them using the install.packages() function.
If the package is on CRAN, we can install it by running the following code in the RStudio Console:
# template for installing a package
install.packages("package_name")
# example of installing a package
install.packages("dplyr")Note that the name of the package needs to be inside quotation marks when using the install.packages() function.
You can run the install.packages() functions within an .R script! However if you choose to do this, please make sure to comment out the line(s) of code that install packages after you have installed those packages. Commenting out the install packages commands will save you time in the future as you will not need to re-install packages each time you run a script.
20.2 Installing a package with pacman (recommended approach)
I highly recommend using the pacman package to install packages instead of using install.packages. You first need to install pacman with install.packages("pacman"), but after that, you should never need to use install.packages. In this course I emphasize p_load() from pacman, which installs the package if necessary and loads it for use in the current R session. This way, you can always keep these commands in your R script without the need to comment out the line after installing the package.
I recommend you begin all your R scripts with the packages that you need as follows. Notice how each package name is on a separate line, which makes it easy to comment out if I don’t need it. Furthermore, each package has a brief comment about it’s utility. Also notice how the package names are not in quotations. This behaviour is different than install.packages which requires the package name to be in quotation marks.
# ------- packages-----------------------------------------------------------
# This script uses the p_load() function from pacman R package,
# which installs if package is absent, and loads for use if already installed
# Ensures the package "pacman" is installed
if (!requireNamespace("pacman")) install.packages("pacman")
pacman::p_load(
# project and file management
#############################
here, # file paths relative to R project root folder
rio, # import/export of many types of data
openxlsx, # import/export of multi-sheet Excel workbooks
# package install and management
################################
remotes, # install from github
# General data management
#########################
tidyverse, # includes many packages for tidy data wrangling and presentation
#dplyr, # data management
#tidyr, # data management
#ggplot2, # data visualization
#stringr, # work with strings and characters
#forcats, # work with factors
#lubridate, # work with dates
#purrr # iteration and working with lists
# statistics
############
janitor, # tables and data cleaning
gtsummary, # making descriptive and statistical tables
broom, # tidy up results from regressions
# plots - general
#################
#ggplot2, # included in tidyverse
cowplot, # combining plots
# patchwork, # combining plots (alternative)
RColorBrewer, # color scales
viridis, # color-blind friendly palettes
# routine reports
#################
knitr,
rmarkdown, # produce PDFs, Word Documents, Powerpoints, and HTML files
# tables for presentation
#########################
flextable, # HTML tables
kableExtra # more customizable knitr::kable
)20.3 Loading a package
Once a package is installed on your computer, you do not have to re-install it in order to use the functions in the package. However, every time you open RStudio and want to use the package, you will need to load the package into your RStudio environment. In this way, R will know where to look for the functions. We can accomplish loading the package into our R environment using the p_load function from the pacman package.
Loading a package into our R environment signals to R that we would like to have access to all the functions available to us in that package. We can load a package, such as the {dplyr} package,126 using the following code:
library(pacman) # load the pacman library to acceess all its functions
# template for loading a package
p_load(package_name)
# example of loading a package
p_load(dplyr)We only have to install a package once, but to use it, we have to load it each time we start a new R session.
There are over 18,000 packages on available on CRAN. It is likely that there are function names that are identical from two different libraries. For example, both plyr and Hmisc provide a summarize() function. If you load plyr, then Hmisc, summarize() will refer to the Hmisc version. But if you load the packages in the opposite order, summarize() will refer to the plyr version. This can be confusing. Instead, you can explicitly refer to specific functions: Hmisc::summarize() and plyr::summarize(). Then the order in which the packages are loaded won’t matter.
The :: operator makes it explicit which package a function is from. You might have noticed in Section 20.2 above, when loading packages, we used:
pacman::p_load()This can be read as “use the p_load function from the pacman library”. I will use this convention when using functions from specific packages. This will avoid any ambiguities.
20.4 The Relationship Between Packages and Functions
Packages are a collection of functions, and most are designed for a specific dataset, field, and/or set of tasks. Functions are individual components within a package and are what we use to interact with our data.
To put it another way, an R user might write a series of functions that they find themselves needing to use repeatedly in a variety of projects. Instead of re-writing (or copying and pasting) the functions each time they need to use them, an R user can collect all of these individual functions inside a package. They can then load the package any time that they want to use the functions, using a single line of code instead of tens to tens of thousands of lines of code.
20.5 How to Find Packages
As you begin your R learning journey, the bulk of the packages you will need to use are either already included when you install R or available on CRAN. CRAN TaskViews (https://cran.r-project.org/web/views/) is one of the best resources for seeing what packages are available and might be relevant to your work. Other great resources to learn about various R packages are through Twitter (following the “#rstats” hashtag) as well as through Google searches. As R has grown in popularity, Google has gotten significantly better at returning R-related results.
20.6 Learning More About a Package
Sometimes when you look up a package, you will be able to identify the function that you need and continue on your way. Other times, you may need (or want!) to learn more about a specific package. Packages on CRAN might come with something called a “vignette”, which is a worked example using various functions from within the package. You can access a package’s vignette(s) on CRAN TaskViews.
Packages do not need to be submitted to CRAN to be used by the public, and many are available directly from their respective developers via GitHub. Package authors may publish vignettes or blog posts about their package, and other R users may also publish tutorials about a specific package. If you find yourself on GitHub looking at information for a package, more often than not, the README file will have good information for getting started with a package.
For example, in Figure 20.1, we show the CRAN homepage for the casebase package.
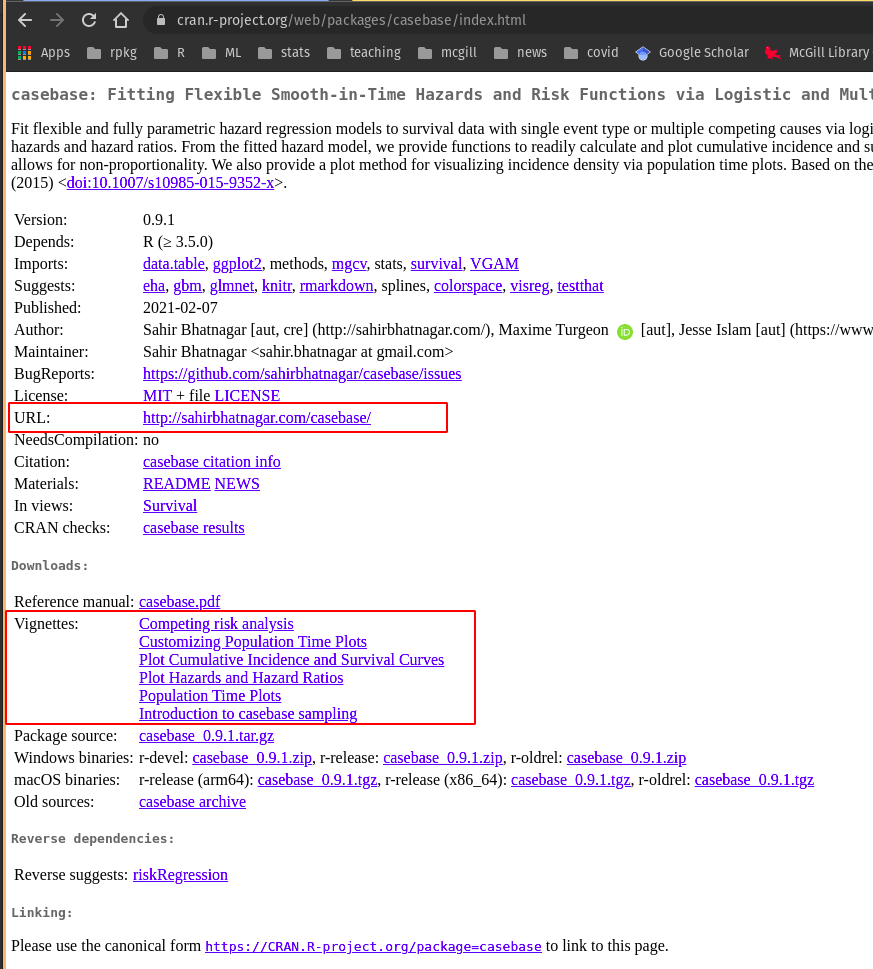
Figure 20.1: CRAN homepage for the casebase package. Much of the information about a package can be found on this landing page. You might also find more information on the package website with can be found in the URL link boxed in red.